sociogenomics2022
Upload the software plink on the folder Software
cd $HOME
cd Sociogenomics/Software
wget https://s3.amazonaws.com/plink1-assets/plink_linux_x86_64_20210606.zip
unzip plink_linux_x86_64_20210606.zip
Alternative command if wget is not installed in your system
curl -o plink_linux_x86_64_20210606.zip https://s3.amazonaws.com/plink1-assets/plink_linux_x86_64_20210606.zip
Chek file permissions
- chmod +rwx filename to add permissions.
- chmod -rwx directoryname to remove permissions.
- chmod +x filename to allow executable permissions.
- chmod -wx filename to take out write and executable permissions.
chmod +x plink
./plink --help
Create symbolic links
cd $HOME/Sociogenomics
ln -s Software/plink
./plink --help
Clean Data and Results folder
` rm Data/. rm Results/. `
UPLOAD FILE week2.zip from Virtuale and unzip files in Data folder
cd Data/
unzip week2.zip
mv week2/*.* ./
rm -r __MACOSX/
rm -r week2
cd $HOME/Sociogenomics
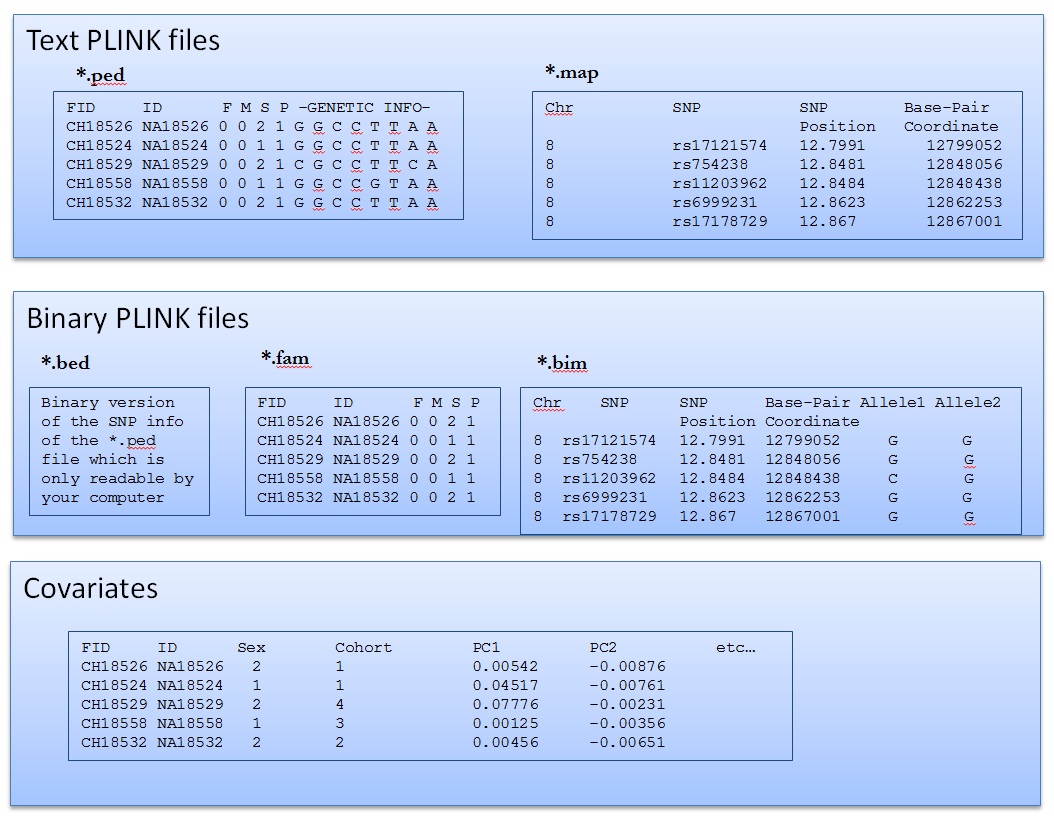
Read Binary Plink file
head Data/hapmap-ceu.bim
head Data/hapmap-ceu.fam
.bed files are not readable!
head Data/hapmap-ceu.bed
Recode into map and ped files
./plink --bfile Data/hapmap-ceu --recode --out Results/hapmap-ceu
head Results/hapmap-ceu.map
head -1 Results/hapmap-ceu.ped
Import VCF into plink
./plink --vcf Data/ALL.chr21.vcf.gz --make-bed --out Results/test_vcf
Select individuals
./plink --bfile Data/hapmap-ceu \
--keep Data/list.txt \
--make-bed --out Results/selectedIndividuals
Select individuals with genotype at least 95% complete
./plink --bfile Data/hapmap-ceu --make-bed --mind 0.05 --out Results/highgeno
Select specific markers
./plink --bfile Data/hapmap-ceu \
--snps rs9930506 \
--make-bed \
--out Results/rs9930506sample
Merge genetic files
./plink --bfile Data/HapMap_founders \
--bmerge HapMap_nonfounders \
--make-bed --out Results/merged_file
Attach a phenotype
head Data/1kg_EU_qc.fam
head Data/1kg_EU_qc.bim
head Data/BMI_pheno.txt
./plink --bfile Data/1kg_EU_qc\
--pheno Data/BMI_pheno.txt \
--make-bed --out Results/1kg_EU_BMI
head Data/1kg_EU_BMI.fam
Descriptive Statistics
Allele frequency
./plink --bfile Data/hapmap-ceu --freq --out Results/Allele_Frequency
head Results/Allele_Frequency.frq
Missing values
individuals
./plink --bfile Data/hapmap-ceu --missing --out Results/missing_data
variants
head Data/missing_data.imiss
Filter females
./plink --bfile Data/hapmap-ceu \
--filter-females \
--make-bed \
--out Results/hapmap_filter_females
Quality control
./plink --bfile Data/1kg_hm3 --mind 0.05 --make-bed --out Results/1kg_hm3_mind005
Calculate heterozygocity
./plink --bfile Data/1kg_hm3 --het --out Results/1kg_hm3_het
Check discordant sex
./plink --bfile Data/hapmap-ceu --check-sex --out Results/hapmap_sexcheck
Low call-rate SNPS
./plink --bfile Data/1kg_hm3 --geno 0.05 --make-bed --out Results/1kg_hm3_geno
Allele frequency
./plink --bfile Data/1kg_hm3 --maf 0.01 --make-bed --out Results/1kg_hm3_maf
deviation from HWE
./plink --bfile Data/1kg_hm3 --hwe 0.00001 --make-bed --out Results/1kg_hm3_hwe
Plink QC
./plink --bfile Data/1kg_hm3 \
--mind 0.03 \
--geno 0.05 \
--maf 0.01 \
--hwe 0.00001 \
--exclude Data/individuals_failQC.txt \
--make-bed --out Results/1kg_hm3_QC